Research into matching patterns of over a half-million AncestryDNA members translates into new DNA matching discoveries
Among over 500,000 AncestryDNA customers, more than 35 million 4th cousin relationships have been identified – a number that continues to grow rapidly at an exponential rate. While that means millions of opportunities for personal discoveries by AncestryDNA members, it also means a lot of data that the AncestryDNA science team can put back into research and development for DNA matching.
At the Institute for Genetic Genealogy Annual Conference in Washington, D.C. this past weekend, I spoke about some of the AncestryDNA science team’s latest exciting discoveries – made by carefully studying patterns of DNA matches in a 500,000-member database.

DNA matching means identifying pairs of individuals whose genetics suggest that they are related through a recent common ancestor. But DNA matching is an evolving science. By analyzing the results from our current method for DNA matching, we have learned how we might be able to improve upon it for the future.
The science team targeted our research of the DNA matching data so that we could obtain insight into two specific steps of the DNA matching procedure.
Remember that a person gets half of their DNA from each of their parents – one full copy from their mother and one from their father. The problem is that your genetic data doesn’t tell us which parts of your DNA you inherited from the same parent. The first step of DNA matching is called phasing, and determines the strings of DNA letters that a person inherited from each of their parents. In other words, phasing distinguishes the two separate copies of a person’s genome.
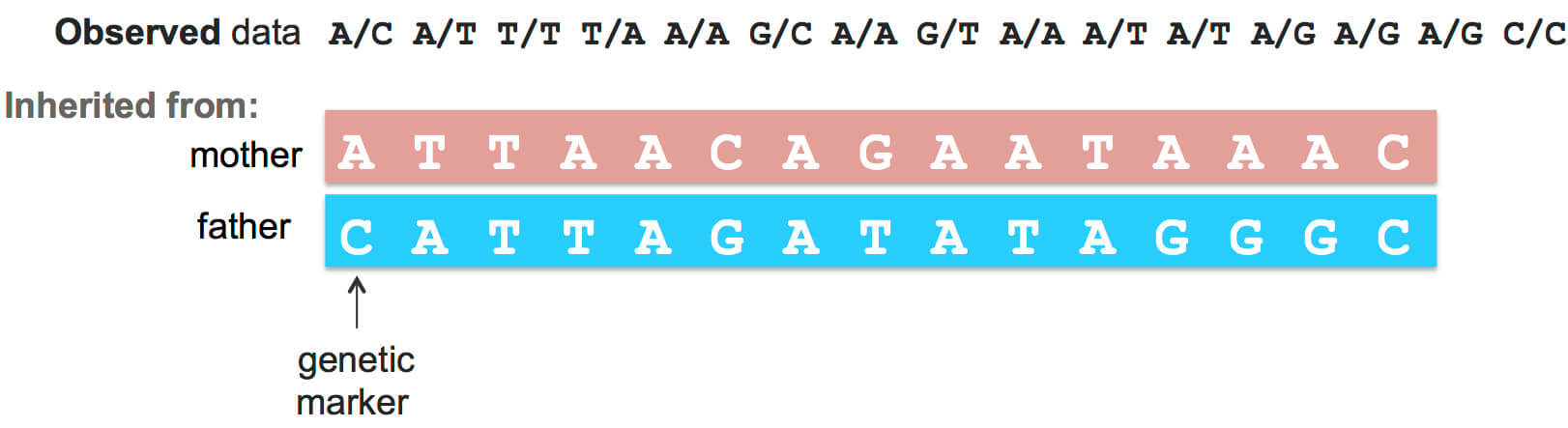
If we had DNA from everyone’s parents, phasing someone’s DNA would be easy. But unfortunately, we don’t. So instead, phasing someone’s DNA is often based on a “reference” dataset of people in the world who are already phased. Typically, those reference sets are rather small (around one thousand people).
Studies of customer data led us to find that we could incorporate data from hundreds of thousands of existing customers into our reference dataset. The result? Phasing that is more accurate, and faster. Applying this new approach would mean a better setup for the next steps of DNA matching.
The second step in DNA matching is to look for pieces of DNA that are identical between individuals. For genealogy research, we’re interested in DNA that’s identical because two people are related from a recent common ancestor. This is called DNA that is identical by descent, or IBD. IBD DNA is what leads to meaningful genealogical discoveries: allowing members to connect with cousins, find new ancestors, and collaborate on research.
But there other reasons why two people’s DNA could be identical. After all, the genomes of any two humans are 99.9% identical. Pieces of DNA could be identical between two people because they are both human, because they are of the same ethnicity, or because they share some other more ancient shared history. We call these pieces of DNA only identical by state (IBS), because the DNA could be identical for a reason other than a recent common ancestor.
We sought to understand the causes of identical pieces of DNA between more than half a million AncestryDNA members. Our in-depth study of these matches led us to find that in certain places of the genome, thousands of people were being estimated to have DNA that was identical to one another.
What we found is that thousands of people all having matching DNA isn’t a signal of all of them being closely related to one another. Instead, it’s likely a hallmark of a more ancient shared history between those thousands of individuals – or IBS.

In other words, our analysis revealed that in a few cases where we thought people’s DNA was identical by descent, it was actually identical by state. These striking matching patterns were only apparent after viewing the massive amount of matching data that we did.
So while the data suggested that our algorithms had room for improvement, that same data gave us the solution. After exploring a large number of potential fixes and alternative algorithms, we discovered that the best way to address the problem was to use the observed DNA matches to determine which were meaningful for genealogy (IBD) – and distinguish them from those due to more ancient shared history. In other words, the matching data itself has the power to help us tease apart the matches that we want to keep from those that we want to throw away.
The AncestryDNA science team’s efforts – poring through mounds and mounds of DNA matches – have paid off. From preliminary testing, it appears that these latest discoveries relating to both steps of DNA matching may lead to dramatic DNA matching improvements. In the future, this may translate to a higher-quality list of matches for each AncestryDNA member: fewer false matches, and a few new matches too.
In addition to the hard work of the AncestryDNA science team, the huge amount of DNA matching data from over a half-million AncestryDNA members is what has enabled these new discoveries. Carefully studying the results from our existing matching algorithms has now allowed us to complete the research and development “life cycle” of DNA matching: translating real data into future advancements in the AncestryDNA experience.